About our work
We use a broad systems approach to integrate genomics, structure and function and evolutionary perspectives to study the eye and its diseases.
Current research
We study the molecular basis for major diseases of the aging eye, particularly age-related macular degeneration (AMD) and cataract, using a wide range of approaches including genomics, cell and structural biology.
AMD and Aging Retina:
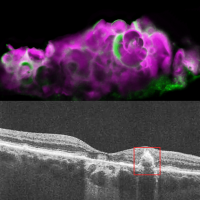
Using a novel model to study cells derived from human retinal pigment epithelium (RPE) we have shown that serum-deprivation induces programmed responses that are highly relevant to AMD. These include upregulation of cholesterol and fatty-acid pathways and deposition of lipids, proteins and minerals that mimic AMD. This model allowed us to discover that serum-deprivation of RPE induces expression of amelotin (AMTN) a protein involved in dental enamel formation and that this is associated with mineralization of calcium phosphate to hydroxyapatite (HAP) in RPE. HAP deposits are markers for disease progression in “dry” AMD, a widespread condition that currently lacks treatments. We have developed a mouse model for human AMTN expression which is being used to test siRNA and drug approaches to blocking AMTN expression or HAP formation. This presents exciting possibilities for new treatments aimed at slowing progression to vision loss in dry AMD.
We are also looking at wider implications for the unexpected role of AMTN in cellular damage and stress responses and we are collaborating on structural studies on AMTN, an intrinsically disordered protein, to gain further functional insights.
Another retina discovery involves retbindin, a retina-specific protein we discovered through the NEIBank genomics project. We have now shown that retbindin has high affinity for the visual chromophore 11-cis retinal and is unexpectedly involved in the visual cycle, particularly related to light sensitivity of dark-adapted photoreceptors. Deletion of retbindin in mouse leads to several aspects of premature aging, including loss of melanopsin expression in light-sensitive retinal ganglion cells. We are using the retbindin knockout mice to develop a more complete model for AMD and retinal aging.
Cataract and Lens:
We have a long-term interest in the structure and function of crystallins and other proteins of the eye lens. In recent work, we used crystallography to capture a remarkable unfolding/aggregation intermediate of a γ-crystallin. This structure shows how well-ordered proteins can undergo domain swapping, lose symmetry and form strained, partially disordered aggregates that can cascade into protein deposition. This shows mechanisms that may occur in cataract and may also be relevant to other diseases involving protein aggregation.
We are collaborating on studies revealing unexpected roles for γS-crystallin in retina, particularly in the aging retina. We are also continuing structural studies, including use of CryoEM to investigate structure function in lengsin, another NEIBank discovery, with an important role in lens organization.
Highlights from the Lab
Selected publications
Sagar, V. and Wistow, G. (2022) Acquired disorder and asymmetry in a domain-swapped model for g-crystallin aggregation. J. Mol. Biol. 434(9):167559.
Fan, J., Rajapakse, D., Peterson, K., Lerner, J., Parsa, S., Ponduri, A., Sagar, V., Duncan, T., Dong, L. and Wistow, G. (2021) Retbindin mediates light-damage in mouse retina while its absence leads to premature retinal aging. Exp Eye Res. 209:108698
Rajapakse, D., Peterson, K., Mishra, S., Fan, J., Lerner, J., Campos, M. and Wistow, G. (2020) Amelotin, a promoter of calcium mineralization, is induced by serum-deprivation in retinal pigment epithelial cells in culture and localizes to hydroxyapatite deposits in dry AMD. Transl. Res. 219:45-62.
Fan, J., Lerner, J., Wyatt, K.K., Cai, P, Peterson, K., Dong, L. and Wistow, G. (2018) The Klotho-related protein Klph (Lctl) has preferred expression in lens and is essential for expression of Clic5 and normal lens suture formation. Exp. Eye Res. 169:111-121.
Rajapakse, D., Peterson, K., Mishra, S. and Wistow G. (2017) Serum starvation of ARPE-19 changes the cellular distribution of cholesterol and Fibulin3 in patterns reminiscent of Age-related Macular Degeneration. Exp. Cell Res. 361(2):333-341.
Molecular Structure and Functional Genomics key staff
| Name | Title | Phone | |
|---|---|---|---|
| Katherine Peterson, Ph.D. | Staff Scientist | Petersonk@nei.nih.gov | 301-402-3452 |
| Graeme J. Wistow, Ph.D. | Senior Investigator | WistowG@NEI.NIH.GOV | 301-402-3452 |
| Jianguo Fan, Ph.D. | Staff Scientist | fanj@nei.nih.gov | 301-402-4812 |
| Dinusha Rajapakse | Research Fellow | dinusha.rajapakse@nih.gov | 301-402-5351 |