
Left: Adult fruit flies normally have compound eyes – essentially ~750 mini eyes, called ommatidia, arranged in a lattice (left). The eyes of flies with mutations in the eyeless gene are either very small or completely missing (right). Reprinted with permission1.
Right: Drosophila chromosomes. The smallest chromosome, number 4 (two dots near the center of the image), contains the eyeless gene. Reprinted with permission4.
A century ago, the fruit fly was making waves. Easy to catch and breed, (many captured at outdoor vegetable stands, and grown on old banana peels or rotting fruit) the humble fruit fly became a popular model organism in the exciting new field of genetics. Long before the discovery of DNA’s structure, researchers were already feverishly mapping physical traits to regions of the fly’s genome.
One such researcher, a young graduate student at Columbia University by the name of Mildred Hoge, isolated a group of flies that exhibited small – or even completely missing – eyes. Upon mapping the affected genetic region to the smallest of the fly’s chromosomes, number 4, she named the mutated strain eyeless, or ey for short. After her first eyeless publication in 19153, her later research revealed key roles for eyeless in the development of the eye and the optic nerve. But it wouldn’t be until the 1990s, long after Hoge’s death, that scientists came to understand the true importance of the eyeless gene.
eyeless, it turns out, is a master regulator for the development of the eye and other structures not just in flies, but throughout the animal kingdom. In mammals like mice and humans, eyeless has a counterpart in a highly similar gene family called PAX6. Mutations that affect the levels of PAX6 protein during development can cause aniridia, a malformation of the iris and other parts of the eye. In more severe cases, problems with PAX6 protein can affect the formation of the brain and other critical structures of the body.
Tools developed and knowledge gained over the past century have made this type of research quicker, but in many ways, small creatures like the fruit fly continue to open critical windows to the nature of life -- and of our own bodies. In the vision field, researchers turn to a variety of small, non-mammalian animal models to help bridge critical gaps in our scientific knowledge.

"An observational science, though important because of its addition to the storehouse of human knowledge, is not vital if it has no practical applications… Building on the firm foundation of observations of early workers, modern biologists have been able to improve both man’s environment and his health. The new knowledge they have acquired has made possible better surroundings and better opportunities for economic life."5
– Mildred Hoge Richards, 1938
Mildred Hoge, 1908. Image courtesy of Gaucher College Special Collections and Archives.
Exploiting a Long History
While the fruit fly’s eye looks nothing like ours, the same key features of the fly that made it a great model organism in 1915 still hold true today. Easy and inexpensive to grow, and with a deep history of research, modern researchers continue to study how the fruit fly develops, asking questions like: What makes an eye an eye? Why do both eyes end up the same size? How does an eye know when to stop growing? What keeps an eye from turning into some other body part?
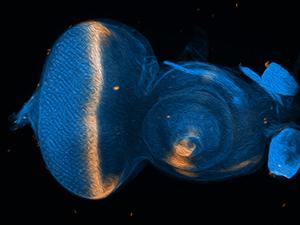
Cell fate changes driven by PAX6 take place while the fly is in the larval part of its lifecycle. The cells that will eventually become structures like the eye, head, leg or wing in the adult fly exist as a collection of small disc-shaped organs within the larva called “imaginal discs”. Image shows an eye-antennal imaginal disc with the gene decapentaplegic (dpp) in peach. Image courtesy of Justin Kumar, Ph.D.
Justin Kumar, Ph.D., a researcher at University of Indiana, Bloomington, is delving deep into questions like these, building on older, 20th-century research using the latest tools and technologies.
Scientists in the early days “had very few instruments in their toolbox, but they were incredibly clever. They would just look and ask, ‘How does this work?’ said Kumar. “Fundamental questions, like ‘How does an eye know how large to grow? When should it start growing? And when should it stop?’ The answers that they came up with were astonishingly accurate. I find myself putting in the details, attaching the molecules to the concepts they had identified a long time ago.”
One key area of interest for Kumar is exploring cell fate – essentially, how a developing tissue becomes set as one kind of tissue and not another, like an eye and not a wing or an antenna.
As Hoge discovered early on, the PAX6 family of proteins, including Eyeless, drive the development of tissues that make up the eye and head. The PAX6 proteins (transcription factors), along with associated genes, work by turning on “eye” genes and turning off “not eye” genes.
Kumar has found that just changing PAX6 on its own doesn’t let a cell become any tissue. The cells are still limited to types of tissues that are close on the developmental tree (like an eye to antenna, rather than eye to leg or wing). A bigger leap up that developmental tree requires additional changes to regulating proteins.
Model Organism: “An organism suitable for studying a specific trait, disease, or phenomenon, due to its short generation time, characterized genome, or similarity to humans.”
--Nature Education
Kumar has found that a type of protein called an epigenetic enzyme, which adjusts how DNA can be accessed inside the cell, works in tandem with PAX6 proteins to drive many cell fate decisions.
To explore this further, Kumar and his research team have developed a fly system that lets them test a “two hit” model, where they adjust two different proteins at once to see how those two proteins are interacting.
One popular way to figure out what a protein is doing in a developing animal is to “knock out” (remove) the gene that makes that protein from the animal’s genome. In the days of Mildred Hoge, this meant isolating and selectively breeding collections of flies with natural mutations in those genes. Today, researchers can use gene editing tools to control precisely where and how they reduce or remove the gene that makes a particular protein. In Kumar’s case, he’s created a system that works on two genes at once within a fly’s developing eye tissue.
When Kumar turns off both the PAX6 protein and the epigenetic enzyme, suddenly eye tissue can turn into all sorts of things – including a wing. By screening through all the newly modified cell’s gene changes, Kumar can tease out what makes a tissue one type or another.
Kumar thinks “there has to be some sort of combinatorial code underneath these PAX6 and epigenetic enzymes. When you knock them out together, it’s this kind of magic bullet, but that’s hidden when you look at them individually. And that makes sense developmentally, because you don’t want an eye to turn into a wing if PAX6 levels get low,” Kumar said. “That’s why this multi-knockout approach is really exciting… there were clues from the old days, but we had no way of putting them together until we came up with this approach.”
The century-old single-knockout approach, looking at the effects of one gene at a time, is the backbone upon which we’ve built much of modern biological science and medicine. But as Kumar notes, sometimes animals have evolved to protect against mistakes in a single gene, so the importance of those genes can be hidden with a single knockout.
Like most basic science research, delving into how multiple genes interact to make an eye form correctly could have any number of benefits down the road. These cell fate pathways not only give rise to the eye during development, but may come into play later in life in disease, healing from injury, and even during tissue regeneration.
The fruit fly research model system allows researchers like Kumar to study how genes drive the development of the eye.
Harnessing Unique Features
Where some animals, like the fly, have features that make them easy to study, others have capabilities that we would like to harness for ourselves. One example of this is the newt’s ability to regenerate body parts. This family of animals can regrow not only limbs, but complex tissues like those in the eye, up to and including nerve connections to the brain.

Newts like Pleurodeles waltl, the Spanish newt, can regenerate the eye’s lens within months. Images courtesy of Katia Del Rio Tsonis, Ph.D.
Researcher Katia Del Rio-Tsonis, Ph.D., of Miami University, Oxford, OH, is studying how the newt can regrow the eye’s lens. Unlike the fruit fly, where there are existing stocks of animals with wide ranging traits from around the world, newts are an emerging research organism, trickier to breed and not widely available. So rather than using many animals to capture the regeneration process, a critical area for Del Rio-Tsonis has been finding a way to be able to watch lens regeneration happening in real time in a living newt, and discover just how the eye organizes the replacement of this complex tissue.
“Our model system is one of the most exciting systems of vertebrate regeneration. The newt can basically regenerate anything,” said Del Rio-Tsonis. “The eye is really a fantastic model too, because it’s easily manipulated.”
When the lens is removed from a newt’s eye (similar in many ways to cataract removal in a human), the immune system revs into gear to clear out any damaged cells and debris. Then, cells at the top of the iris transform into lens cells, and eventually grow to become a new lens. Over time, the lens grows in size, eventually detaching from the iris and reattaching to the normal connecting tissue (called zonules).
This past year, Del Rio-Tsonis’ research group, in collaboration with the laboratory of Hui Wang, Ph.D., also at Miami University, has found a way to use optical coherence tomography (OCT) to visualize newt lens regeneration in 3D. OCT is a type of imaging often used in humans to diagnose disease – it can scan and reveal parts of the eye at high resolution non-invasively, showing structures like the iris, lens, and the retina (the light-sensitive tissue at the back of the eye).
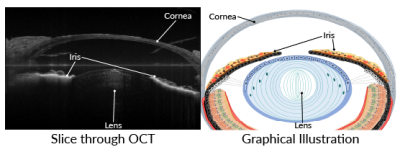
Left: Slice through an OCT scan of an intact newt eye, showing cornea, lens, and iris. Right: Graphical illustration showing the same anatomical structures. Images courtesy of George Tsissios and Katia Del Rio-Tsonis, Ph.D.2
Traditionally, the structure of the lens has been investigated using histology, which involves removing the tissue and viewing it under a microscope.
“We’ve been wanting to find a better way to assess the lens regeneration process, rather than regular histology, which is time consuming and uses many more animals. I’m super excited because now we can image when the lens is intact, and then we can remove it and you follow the lens regeneration process in the same animal,” Del Rio-Tsonis said. “With OCT, you can see basically everything, even the injury to the cornea from surgery, as well as the re-formed zonule fibers. We can now test treatments to inhibit or induce lens regeneration and follow the process in individual animals.”
Seeing the regeneration of a lens play out in real time gives scientists a view into a complex, fascinating dance of cells and tissues reorganizing themselves to rebuild a part of an existing eye. Not only does this reduce the number of animals required to undertake a study of lens regeneration, but viewing the entire process in individual animals can yield unexpected findings as well.
“Along with other imaging techniques, OCT can help us answer a lot of questions about the role of the immune system and blood vessels in regeneration, as well as how the lens connects to the zonule fibers. We can also screen treatments and drugs that affect lens regeneration so much faster now. And there are features we did not know that we discovered just from the 3D OCT, like the shape of the lens vesicle,” said Del Rio-Tsonis. “You can actually follow the 3D shape over time. It’s beautiful.”
The newt can completely regenerate their eye's lens. Researcher Del Rio-Tsonis studies newt lens regeneration as a way to understand how complex tissues like the eye can regenerate in an adult animal. Newt studies and imaging by George Tsissios and Weihao Chen, Miami University.
Clues for Treating Disease
Newts aren’t the only animals with regenerative abilities that surpass ours.
Like the newt, the tiny zebrafish is able to regrow parts of the eye, including neurons in the eye’s light sensitive tissue. And this is especially interesting for researchers looking to treat some of the most common blinding conditions in people: age-related macular degeneration (AMD) and glaucoma.
In AMD, the support cells for the eye’s retina can fail, eventually leading to the death of light-sensing photoreceptor cells. In glaucoma, the retinal ganglion cells, nerves that connect the eye to the brain, begin to die off. In humans, neither photoreceptors nor retinal ganglion cells can be replaced, so loss of these cells can lead to permanent blindness.
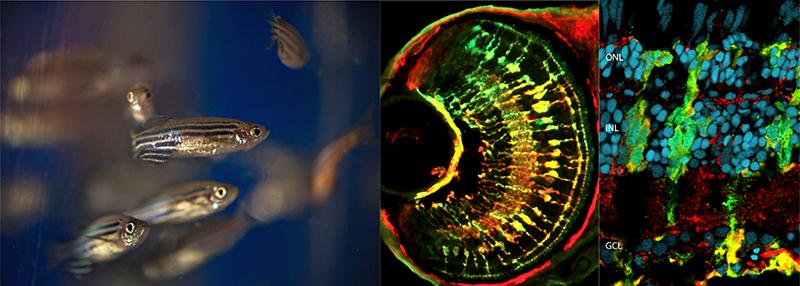
Left: Adult zebrafish. Center: Retinal Müller glia in zebrafish. Right: Injured retina in an adult zebrafish. Images courtesy of Daniel Goldman, Ph.D.
Daniel Goldman, Ph.D., University of Michigan, believes the zebrafish eye holds answers that may one day help us regrow these retinal neurons, both photoreceptors and retinal ganglion cells. When a zebrafish’s retina is damaged, immune cells and special cells called Müller glia activate – this much is similar to what happens in humans. But in humans, the immune response then drives the formation of scar tissue, which has no sensitivity in light, leading to blind areas in the retina. In zebrafish, however, the immune system fades away, and Müller glia dial back their own developmental program, returning to an earlier developmental state. Once there, the zebrafish Müller glia can begin producing new progenitor cells. Those progenitor cells can eventually develop into fresh photoreceptors or retinal ganglion cells, and replace the nerves that have been damaged.
Not only do zebrafish regrow retinal ganglion cells in the retina, they can even regrow the nerve axons that connect the eye to the brain through the optic nerve. After an optic nerve injury, “[the fish] regain sight, it’s really amazing, and it happens really fast. Over a 2-week period, they’ll reconnect with the brain, and the fish can see again,” Goldman said.
Goldman has been working to uncover which genes and which molecular pathways help support the Müller glia’s regenerative capacity.
“There’s a large number of significant differences between the mammalian Müller glia and the zebrafish Müller glia. There are a lot of genes unique to the fish, some of which have gained new functions. Are they some of the key players that are allowing this regenerative response to occur?” Goldman asked. “What we’re trying to do is make it so that Müller glia in the mammal are more fish-like.”
Animals and the Clinic
For most vision researchers, non-mammalian animals are useful to understand the basic biological processes underlying sight. But for clinician scientists like Robert Hufnagel, M.D., Ph.D., director of NEI’s Ophthalmic Genomics Laboratory and chief of the Medical Genetics and Ophthalmic Genomics Unit, these animals can provide a way to understand what’s happening for specific patients who come to the clinic.
“For rare diseases in humans, it can be really difficult to find enough patients with similar clinical diseases and genetic changes to translate those diseases to genotype:phenotype studies in an animal model. Being able to create relevant animal models to help study these rare diseases is important to our patients,” said Hufnagel. Genotype:phenotype studies reveal how individual sections of DNA physically manifest in the body.

Wiegmann's striped gecko (left) and the yellow-headed dwarf gecko (right). Image credit: Andrew Wegerski, Medical Genetics and Ophthalmic Genomics Unit, NEI.
Although he also works with mice and zebrafish, Hufnagel has chosen the gecko as his newest animal model. One of the reasons he chose the gecko is that the animal’s eyes have features that nicely mimic the human eye. Many of the most common animal models – like mice or rats – have what’s known as “rod-dominant” eyes, meaning they see mostly in black and white. They also tend to lack a thin part of the retina called the fovea, which helps humans have high-acuity central vision. The gecko, on the other hand, has a fovea, just like us.
“There are a lot of similarities between the gecko’s eye structure and ours – it shows a very similar pattern of development to the human as well. Right now, we’re doing developmental staging on the gecko’s eye, and testing tools for genetic reprogramming.”
Hufnagel’s plan hinges on a new technology known as “CRISPR”. This technique allows researchers to simply and quickly mutate a particular gene in an animal. Hufnagel plans to use this technique to match geckos to the genetic changes present in his patients. Then he can test novel treatments and therapies in the geckos, with a pretty good idea of what to expect from his patients.
This type of gene editing has worked before in fish and other lizards. “When scientists knock out the gene for tyrosinase, for example, the cause of human oculo-cutaneous albinism, we can evaluate an animal with reduced pigment that resembles the human disease,” Hufnagel said.
In humans, oculo-cutaneous albinism leads not only to reduced melanin pigment in the skin and eye, but also to developmental defects in the fovea, which can cause low vision. By studying this disease in a species like the gecko, which has a fovea, Hufnagel will be able to better understand what pathways are involved in foveal development.
“Not only do new tools like genome editing and genome assembly allow us to do these kinds of manipulations in these animals, it accelerates our ability to choose an animal model based on the features of the organism, rather than the animals that are already available. It gives us new ways to understand the particulars of certain diseases,” Hufnagel said. “As we see patients in clinic and find new genetic diseases, depending on the features we observe in the patient, we can pick the right animal to study their disease.”
References
- Bürgy-Roukala E, Miellet S, Mishra A, Sprecher S. (2013). Early Eye Development: Specification and Determination. In: Singh A, Kango-Singh M (eds). Molecular Genetics of Axial Patterning, Growth and Disease in the Drosophila Eye. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-8232-1_1
- Chen W, Tsissios G, et al. In Vivo Imaging of Newt Lens Regeneration: Novel Insights Into the Regeneration Process. Transl Vis Sci Technol. 2021 Aug 12;10(10):4. doi: 10.1167/tvst.10.10.4. PMID: 34383878
- Hoge, Mildred A. Another Gene in the Fourth Chromosome of Drosophila. The American Naturalist. 1915 49(577):47–49. https://www.jstor.org/stable/2456099
- Kaufman BP. Somatic mitoses of Drosophila melanogaster. J Morphology. 1934 56(1):125-155. https://doi.org/10.1002/jmor.1050560107
- Richards MH. The Changing Place of Biology in World Affairs. Bios. 1938 9(4), 165–175. http://www.jstor.org/stable/4604347